| Title: | Utility Functions to Support and Extend the 'rbmi' Package |
| Version: | 0.3.0 |
| Date: | 2026-02-14 |
| Maintainer: | Mark Baillie <bailliem@gmail.com> |
| Description: | Provides utility functions that extend the capabilities of the reference-based multiple imputation package 'rbmi'. It supports clinical trial analysis workflows with functions for managing imputed datasets, applying analysis methods across imputations, and tidying results for reporting. |
| License: | GPL (≥ 3) |
| Encoding: | UTF-8 |
| RoxygenNote: | 7.3.2 |
| Suggests: | cards, ggplot2, gt, knitr, patchwork, readr, rmarkdown, rstan, spelling, testthat (≥ 3.0.0), tibble |
| Config/testthat/edition: | 3 |
| Language: | en-US |
| Imports: | assertthat, beeca, cli (≥ 3.6.0), dplyr, lifecycle (≥ 1.0.4), purrr, rbmi (≥ 1.4), rlang, tidyr |
| VignetteBuilder: | knitr |
| Depends: | R (≥ 4.1) |
| LazyData: | true |
| URL: | https://github.com/openpharma/rbmiUtils |
| BugReports: | https://github.com/openpharma/rbmiUtils/issues |
| NeedsCompilation: | no |
| Packaged: | 2026-02-16 21:09:40 UTC; bailliem |
| Author: | Mark Baillie |
| Repository: | CRAN |
| Date/Publication: | 2026-02-16 21:40:02 UTC |
rbmiUtils: Utility Functions to Support and Extend the 'rbmi' Package
Description

Provides utility functions that extend the capabilities of the reference-based multiple imputation package 'rbmi'. It supports clinical trial analysis workflows with functions for managing imputed datasets, applying analysis methods across imputations, and tidying results for reporting.
Author(s)
Maintainer: Mark Baillie bailliem@gmail.com (ORCID) [copyright holder]
Authors:
Tobias Muetze tobias.muetze@novartis.com (ORCID)
Jack Talboys jack.talboys@novartis.com
Other contributors:
Lukas A. Widmer (ORCID) [contributor]
See Also
Useful links:
Report bugs at https://github.com/openpharma/rbmiUtils/issues
Example efficacy trial dataset
Description
A simplified example of a simulated trial dataset, with missing data.
Usage
ADEFF
Format
ADEFF
A data frame with 1,000 rows and 10 columns:
- USUBJID
Unique subject identifier
- AVAL
Primary outcome variable
- TRT01P
Planned treatment
- STRATA
Stratification at randomisation
- REGION
Stratification by region
- REGIONC
Stratification by region, numeric code
- BASE
Baseline value of primary outcome variable
- CHG
Change from baseline
- AVISIT
Visit number
- PARAM
Analysis parameter name
Example multiple imputation trial dataset
Description
A simplified example of a simulated trial ADMI dataset
Usage
ADMI
Format
ADMI
A data frame with 100,000 rows and 12 columns:
- USUBJID
Unique patient identifier
- STRATA
Stratification at randomisation
- REGION
Stratification by region
- REGIONC
Stratification by region, numeric code
- TRT
Planned treatment
- BASE
Baseline value of primary outcome variable
- CHG
Change from baseline
- AVISIT
Visit number
- IMPID
Imputation number identifier
- CRIT1FLN
Responder criteria (binary)
- CRIT1FL
Responder criteria (categorical)
- CRIT
Responder criteria (definition)
Apply Analysis Function to Multiple Imputed Datasets
Description
This function applies an analysis function (e.g., ANCOVA) to imputed datasets and stores the results for later pooling. It is designed to work with multiple imputed datasets and apply a given analysis function to each imputation iteration.
Usage
analyse_mi_data(
data = NULL,
vars = NULL,
method = NULL,
fun = rbmi::ancova,
delta = NULL,
...
)
Arguments
data |
A data frame containing the imputed datasets. The data frame should include a variable (e.g., |
vars |
A list specifying key variables used in the analysis (e.g., |
method |
A method object specifying the imputation method used (e.g., Bayesian imputation).
Created using |
fun |
A function that will be applied to each imputed dataset. Defaults to rbmi::ancova.
Other options include |
delta |
A |
... |
Additional arguments passed to the analysis function |
Details
The function loops through distinct imputation datasets (identified by IMPID), applies the provided analysis function fun, and stores the results for later pooling. If a delta dataset is provided, it will be merged with the imputed data to apply the specified delta adjustment before analysis.
Workflow:
Prepare imputed data using
get_imputed_data()orexpand_imputed_data()Define variables using
rbmi::set_vars()Call
analyse_mi_data()to apply analysis to each imputationPool results using
rbmi::pool()Tidy results using
tidy_pool_obj()
Value
An object of class analysis containing the results from applying the analysis function to each imputed dataset.
Pass this to rbmi::pool() to obtain pooled estimates.
See Also
-
rbmi::analyse()which this function wraps -
rbmi::pool()for pooling the analysis results -
tidy_pool_obj()to format pooled results for publication -
get_imputed_data()to extract imputed datasets from rbmi objects -
expand_imputed_data()to reconstruct full imputed data from reduced form -
gcomp_responder_multi()for binary outcome analysis -
validate_data()to check data before imputation
Examples
# Example usage with an ANCOVA function
library(dplyr)
library(rbmi)
library(rbmiUtils)
set.seed(123)
data("ADMI")
# Convert key columns to factors
ADMI$TRT <- factor(ADMI$TRT, levels = c("Placebo", "Drug A"))
ADMI$USUBJID <- factor(ADMI$USUBJID)
ADMI$AVISIT <- factor(ADMI$AVISIT)
# Define key variables for ANCOVA analysis
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CHG",
covariates = c("BASE", "STRATA", "REGION") # Covariates for adjustment
)
# Specify the imputation method (Bayesian) - need for pool step
method <- rbmi::method_bayes(
n_samples = 20,
control = rbmi::control_bayes(
warmup = 20,
thin = 1
)
)
# Perform ANCOVA Analysis on Each Imputed Dataset
ana_obj_ancova <- analyse_mi_data(
data = ADMI,
vars = vars,
method = method,
fun = ancova, # Apply ANCOVA
delta = NULL # No sensitivity analysis adjustment
)
Construct an rbmi analysis object
Description
This is a helper function to create an analysis object that stores the results from multiple imputation analyses. It validates the results and ensures proper class assignment.
This is a modification of the rbmi::as_analysis function. This function is deprecated and will be removed in a future version.
Usage
as_analysis2(results, method, delta = NULL, fun = NULL, fun_name = NULL)
Arguments
results |
A list containing the analysis results for each imputation. |
method |
The method object used for the imputation. |
delta |
Optional. A delta dataset used for adjustment. |
fun |
The analysis function that was used. |
fun_name |
The name of the analysis function (used for printing). |
Value
An object of class analysis with the results and associated metadata.
Convert character variable list to formula
Description
This function is deprecated and will be removed in a future version.
Usage
as_simple_formula2(outcome, covars)
Arguments
outcome |
Character string, the outcome variable. |
covars |
Character vector of covariates. |
Value
A formula object.
Combine Results Across Multiple Analyses
Description
Combines tidy result tibbles from multiple analyses (e.g., different endpoints or subgroups) into a single table with an identifying column.
Usage
combine_results(..., results_list = NULL, id_col = "analysis")
Arguments
... |
Named arguments where each is a tidy result tibble from |
results_list |
Alternative to |
id_col |
Character string specifying the name of the identifier column. Default is "analysis". |
Value
A tibble with all results combined, with an additional column identifying the source analysis.
See Also
-
tidy_pool_obj()to create tidy results from pooled objects -
format_results()to format combined results for reporting
Examples
library(rbmi)
library(dplyr)
# Assuming you have multiple pooled results
# results_week24 <- tidy_pool_obj(pool_obj_week24)
# results_week48 <- tidy_pool_obj(pool_obj_week48)
# Combine them
# combined <- combine_results(
# "Week 24" = results_week24,
# "Week 48" = results_week48
# )
Create IMPID Column for Imputed Datasets
Description
Adds an IMPID column to a list of imputed datasets, converting them to a
single stacked data.frame suitable for use with analyse_mi_data().
Usage
create_impid(imputed_list, id_prefix = "")
Arguments
imputed_list |
A list of data.frames, where each element represents one imputed dataset. |
id_prefix |
Optional character prefix for IMPID values. Default is empty string. |
Details
This function is useful when you have imputed datasets from a source other than rbmi (e.g., from mice or another MI package) and want to use them with rbmiUtils analysis functions.
Value
A single data.frame with all imputed datasets stacked, with an IMPID
column identifying the source imputation.
See Also
-
get_imputed_data()to extract imputed data from rbmi objects -
analyse_mi_data()to analyse stacked imputed data
Examples
# Create example imputed datasets
imp1 <- data.frame(USUBJID = c("S1", "S2"), CHG = c(1.5, 2.5))
imp2 <- data.frame(USUBJID = c("S1", "S2"), CHG = c(1.8, 2.2))
imp3 <- data.frame(USUBJID = c("S1", "S2"), CHG = c(1.6, 2.4))
# Stack with IMPID
stacked <- create_impid(list(imp1, imp2, imp3))
print(stacked)
Describe an rbmi Draws Object
Description
Extracts structured metadata from an rbmi draws object, including method,
formula, sample count, failures, covariance structure, and (for Bayesian
methods) MCMC convergence diagnostics. Returns an S3 object with an
informative print() method.
Usage
describe_draws(draws_obj)
Arguments
draws_obj |
A |
Details
For conditional mean methods, the sample count is displayed as "1 + N" matching the rbmi convention where the first sample is the primary (full-data) fit and the remaining N are jackknife or bootstrap resamples.
For Bayesian methods, MCMC convergence diagnostics (ESS, Rhat) are extracted
from the stanfit object when rstan is available. The converged flag
uses the Rhat < 1.1 threshold matching rbmi's own convention.
Value
An S3 object of class c("describe_draws", "list") containing:
- method
Human-readable method name (e.g., "Bayesian (MCMC via Stan)")
- method_class
Raw class name: "bayes", "approxbayes", or "condmean"
- n_samples
Total number of samples
- n_failures
Number of failed samples
- formula
Deparsed model formula string
- covariance
Covariance structure (e.g., "us")
- same_cov
Logical; whether same covariance is used across groups
- condmean_type
(condmean only) "jackknife" or "bootstrap"
- n_primary
(condmean only) Always 1
- n_resampled
(condmean only) Number of resampled draws
- bayes_control
(bayes only) List with warmup, thin, chains, seed
- mcmc
(bayes with stanfit only) List with rhat, ess, max_rhat, min_ess, n_params, converged
See Also
-
rbmi::draws()to create draws objects -
rbmi::method_condmean(),rbmi::method_bayes(),rbmi::method_approxbayes()for method specification
Examples
## Not run:
library(rbmi)
library(dplyr)
data("ADEFF", package = "rbmiUtils")
# Prepare ADEFF data for rbmi pipeline
ADEFF <- ADEFF |>
mutate(
TRT = factor(TRT01P, levels = c("Placebo", "Drug A")),
USUBJID = factor(USUBJID),
AVISIT = factor(AVISIT, levels = c("Week 24", "Week 48"))
)
vars <- set_vars(
subjid = "USUBJID", visit = "AVISIT", group = "TRT",
outcome = "CHG", covariates = c("BASE", "STRATA", "REGION")
)
dat <- ADEFF |> select(USUBJID, STRATA, REGION, TRT, BASE, CHG, AVISIT)
draws_obj <- draws(
data = dat, vars = vars,
method = method_bayes(n_samples = 100)
)
# Inspect the draws object
desc <- describe_draws(draws_obj)
print(desc)
# Programmatic access to metadata
desc$method
desc$n_samples
desc$formula
## End(Not run)
Describe an rbmi Imputation Object
Description
Extracts structured metadata from an rbmi imputation object, including
method, number of imputations (M), reference arm mappings, subject count,
and a missingness breakdown by visit and treatment arm. Returns an S3 object
with an informative print() method.
Usage
describe_imputation(impute_obj)
Arguments
impute_obj |
An |
Details
The missingness table is built by cross-tabulating
impute_obj$data$is_missing by visit and treatment group. Each row shows
the total number of subjects in that group, how many had missing data at
that visit, and the percentage missing.
Value
An S3 object of class c("describe_imputation", "list") containing:
- method
Human-readable method name (e.g., "Bayesian (MCMC via Stan)")
- method_class
Raw class name: "bayes", "approxbayes", or "condmean"
- n_imputations
Number of imputations (M)
- references
Named character vector of reference arm mappings, or NULL
- n_subjects
Total number of unique subjects
- visits
Character vector of visit names
- missingness
A data.frame with columns: visit, group, n_total, n_miss, pct_miss
See Also
-
rbmi::impute()to create imputation objects -
describe_draws()for inspecting draws objects
Examples
## Not run:
library(rbmi)
library(dplyr)
data("ADEFF", package = "rbmiUtils")
ADEFF <- ADEFF |>
mutate(
TRT = factor(TRT01P, levels = c("Placebo", "Drug A")),
USUBJID = factor(USUBJID),
AVISIT = factor(AVISIT, levels = c("Week 24", "Week 48"))
)
vars <- set_vars(
subjid = "USUBJID", visit = "AVISIT", group = "TRT",
outcome = "CHG", covariates = c("BASE", "STRATA", "REGION")
)
dat <- ADEFF |> select(USUBJID, STRATA, REGION, TRT, BASE, CHG, AVISIT)
draws_obj <- draws(
data = dat, vars = vars,
method = method_bayes(n_samples = 100)
)
impute_obj <- impute(
draws_obj,
references = c("Placebo" = "Placebo", "Drug A" = "Placebo")
)
# Inspect the imputation
desc <- describe_imputation(impute_obj)
print(desc)
# Programmatic access
desc$n_imputations
desc$missingness
desc$references
## End(Not run)
Create Regulatory-Style Efficacy Summary Table
Description
Takes an rbmi pool object and produces a publication-ready gt table in the style of CDISC/ICH Table 14.2.x. The table displays least squares means by treatment arm, treatment differences, confidence intervals, and p-values, organized by visit row groups.
Usage
efficacy_table(
pool_obj,
title = NULL,
subtitle = NULL,
digits = 2,
ci_level = NULL,
arm_labels = NULL,
pval_digits = 3,
pval_threshold = 0.001,
font_family = NULL,
font_size = NULL,
row_padding = NULL,
...
)
Arguments
pool_obj |
A pooled analysis object of class |
title |
Optional character string for the table title. |
subtitle |
Optional character string for the table subtitle. |
digits |
Integer. Number of decimal places for estimates and standard errors. Default is 2. |
ci_level |
Numeric. Confidence level for CI column labeling. If |
arm_labels |
Named character vector with elements |
pval_digits |
Integer. Number of decimal places for p-values. Default is 3. |
pval_threshold |
Numeric. P-values below this threshold are displayed as "< threshold". Default is 0.001. |
font_family |
Optional character string specifying the font family for
the table. When |
font_size |
Optional numeric value specifying the table font size in
pixels. When |
row_padding |
Optional numeric value specifying the vertical padding
for data rows in pixels. When |
... |
Additional arguments passed to |
Details
This function assumes a single-parameter-per-visit pool object (the standard
output from an rbmi ANCOVA or MMRM pipeline). It internally calls
tidy_pool_obj() to parse the pool object, then constructs the gt table.
Arm labels: Use the arm_labels parameter to customize arm names in the
table. For example, arm_labels = c(ref = "Placebo", alt = "Drug A") will
display "LS Mean (Placebo)" and "LS Mean (Drug A)" instead of the defaults.
Customization: The returned gt object can be further customized using
standard gt piping, e.g., efficacy_table(pool_obj) |> gt::tab_options(...).
Example output:
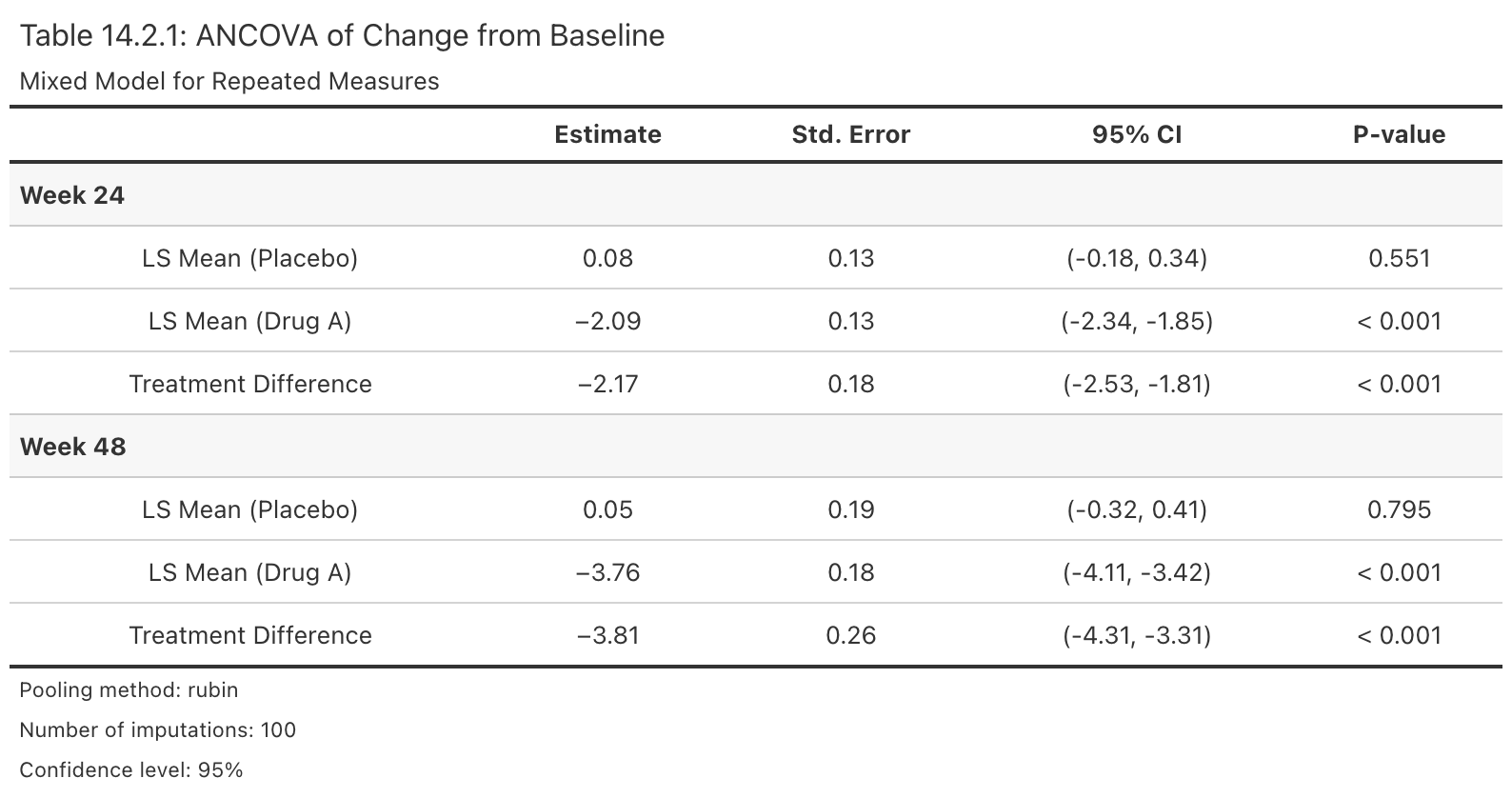
Value
A gt table object of class gt_tbl.
See Also
-
tidy_pool_obj()for the underlying data transformation -
format_pvalue()for p-value formatting rules -
rbmi::pool()to create pool objects
Examples
if (requireNamespace("gt", quietly = TRUE)) {
library(rbmi)
data("ADMI", package = "rbmiUtils")
ADMI$TRT <- factor(ADMI$TRT, levels = c("Placebo", "Drug A"))
ADMI$USUBJID <- factor(ADMI$USUBJID)
ADMI$AVISIT <- factor(ADMI$AVISIT)
vars <- set_vars(
subjid = "USUBJID", visit = "AVISIT", group = "TRT",
outcome = "CHG", covariates = c("BASE", "STRATA", "REGION")
)
method <- method_bayes(
n_samples = 20,
control = control_bayes(warmup = 20, thin = 1)
)
ana_obj <- analyse_mi_data(ADMI, vars, method, fun = ancova)
pool_obj <- pool(ana_obj)
# Basic table
tbl <- efficacy_table(pool_obj)
# Publication-styled table
efficacy_table(
pool_obj,
title = "Table 14.2.1: ANCOVA of Change from Baseline",
subtitle = "Mixed Model for Repeated Measures",
arm_labels = c(ref = "Placebo", alt = "Drug A"),
font_size = 12,
row_padding = 4
)
}
Expand Reduced Imputed Data to Full Dataset
Description
Reconstructs the full imputed dataset from a reduced form by merging imputed
values back with the original observed data. This is the inverse operation of
reduce_imputed_data().
Usage
expand_imputed_data(reduced_data, original_data, vars)
Arguments
reduced_data |
A data.frame containing only the imputed values, as
returned by |
original_data |
A data.frame containing the original dataset before imputation, with missing values in the outcome column. |
vars |
A |
Details
For each imputation (identified by IMPID), this function:
Starts with the original data (observed values)
Replaces missing outcome values with the corresponding imputed values
Stacks all imputations together
Value
A data.frame containing the full imputed dataset with one complete
dataset per IMPID value. The structure matches the output of
get_imputed_data().
See Also
-
rbmi::impute()which creates the imputed datasets this function operates on -
reduce_imputed_data()to create the reduced dataset -
get_imputed_data()to extract imputed data from an rbmi imputation object
Examples
library(rbmi)
library(dplyr)
# Example with package data
data("ADMI", package = "rbmiUtils")
data("ADEFF", package = "rbmiUtils")
# Prepare original data to match ADMI structure
original <- ADEFF |>
mutate(
TRT = TRT01P,
USUBJID = as.character(USUBJID)
)
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CHG"
)
# Reduce and then expand
reduced <- reduce_imputed_data(ADMI, original, vars)
expanded <- expand_imputed_data(reduced, original, vars)
# Verify expansion
cat("Original ADMI rows:", nrow(ADMI), "\n")
cat("Expanded rows:", nrow(expanded), "\n")
Extract variable names from model terms
Description
Takes a character vector including potentially model terms like * and : and
extracts out the individual variables.
This function is deprecated and will be removed in a future version.
Usage
extract_covariates2(x)
Arguments
x |
A character vector of model terms. |
Value
A character vector of unique variable names.
Extract Least Squares Means
Description
Convenience function to extract only least squares mean estimates from tidy results.
Usage
extract_lsm(results, visit = NULL, arm = NULL)
Arguments
results |
A tibble from |
visit |
Optional character vector of visits to filter. If NULL (default), returns results for all visits. |
arm |
Optional character: "ref" for reference arm, "alt" for alternative arm, or NULL (default) for both. |
Value
A tibble containing only LSM rows.
See Also
-
tidy_pool_obj()to create tidy results -
extract_trt_effects()to extract treatment effects
Examples
library(rbmi)
# Assuming you have a tidy result
# tidy_result <- tidy_pool_obj(pool_obj)
# all_lsm <- extract_lsm(tidy_result)
# ref_lsm <- extract_lsm(tidy_result, arm = "ref")
Extract Treatment Effect Estimates
Description
Convenience function to extract only treatment comparison estimates from tidy results, filtering out least squares means.
Usage
extract_trt_effects(results, visit = NULL)
Arguments
results |
A tibble from |
visit |
Optional character vector of visits to filter. If NULL (default), returns results for all visits. |
Value
A tibble containing only treatment effect rows.
See Also
-
tidy_pool_obj()to create tidy results -
extract_lsm()to extract least squares means
Examples
library(rbmi)
# Assuming you have a tidy result
# tidy_result <- tidy_pool_obj(pool_obj)
# trt_effects <- extract_trt_effects(tidy_result)
# trt_week24 <- extract_trt_effects(tidy_result, visit = "Week 24")
Format Estimate with Confidence Interval
Description
Formats a point estimate with its confidence interval in standard publication format: "estimate (lower, upper)".
Usage
format_estimate(estimate, lower, upper, digits = 2, sep = ", ")
Arguments
estimate |
Numeric vector of point estimates. |
lower |
Numeric vector of lower confidence interval bounds. |
upper |
Numeric vector of upper confidence interval bounds. |
digits |
Integer. Number of decimal places for rounding. Default is 2. |
sep |
Character. Separator between lower and upper bounds. Default is ", ". |
Details
The function formats estimates as "X.XX (X.XX, X.XX)" by default. All three
input vectors must have the same length. NA values in any position result
in NA_character_ for that element.
Value
A character vector of formatted estimates with confidence intervals.
Examples
# Single estimate
format_estimate(1.5, 0.8, 2.2)
#> "1.50 (0.80, 2.20)"
# Multiple estimates
format_estimate(
estimate = c(-2.5, -1.8),
lower = c(-4.0, -3.2),
upper = c(-1.0, -0.4)
)
#> "-2.50 (-4.00, -1.00)" "-1.80 (-3.20, -0.40)"
# More decimal places
format_estimate(0.234, 0.123, 0.345, digits = 3)
#> "0.234 (0.123, 0.345)"
# Different separator
format_estimate(1.5, 0.8, 2.2, sep = " to ")
#> "1.50 (0.80 to 2.20)"
Format P-values for Publication
Description
Formats p-values according to common publication standards, with configurable thresholds and decimal places.
Usage
format_pvalue(x, digits = 3, threshold = 0.001, html = FALSE)
Arguments
x |
A numeric vector of p-values. |
digits |
Integer. Number of decimal places for rounding. Default is 3. |
threshold |
Numeric. P-values below this threshold are displayed as "< threshold". Default is 0.001. |
html |
Logical. If |
Details
The function applies the following rules:
P-values below
thresholdare formatted as "< 0.001" (or HTML equivalent)P-values >=
thresholdare rounded todigitsdecimal places-
NAvalues are preserved asNA_character_ Values > 1 or < 0 return
NA_character_with a warning
Value
A character vector of formatted p-values.
Examples
# Basic usage
format_pvalue(0.0234)
#> "0.023"
format_pvalue(0.00005)
#> "< 0.001"
# Vector input
pvals <- c(0.5, 0.05, 0.001, 0.0001, NA)
format_pvalue(pvals)
#> "0.500" "0.050" "0.001" "< 0.001" NA
# Custom threshold
format_pvalue(0.005, threshold = 0.01)
#> "< 0.01"
# HTML output
format_pvalue(0.0001, html = TRUE)
#> "< 0.001"
Format Results for Reporting
Description
Formats a tidy results tibble for publication-ready reporting, with options for rounding, confidence interval formatting, and column selection.
Usage
format_results(
results,
digits = 2,
ci_format = c("parens", "brackets", "dash"),
pval_digits = 3,
include_se = FALSE
)
Arguments
results |
A tibble from |
digits |
Integer specifying the number of decimal places for estimates. Default is 2. |
ci_format |
Character string specifying CI format. Options are: "parens" for "(LCI, UCI)", "brackets" for "\[LCI, UCI\]", or "dash" for "LCI - UCI". Default is "parens". |
pval_digits |
Integer specifying decimal places for p-values. Default is 3. |
include_se |
Logical indicating whether to include standard error column. Default is FALSE. |
Value
A tibble with formatted columns suitable for reporting.
See Also
-
tidy_pool_obj()to create tidy results -
combine_results()to combine multiple analyses
Examples
library(rbmi)
# Assuming you have a tidy result
# tidy_result <- tidy_pool_obj(pool_obj)
# formatted <- format_results(tidy_result, digits = 3, ci_format = "brackets")
Format Results Table for Publication
Description
Adds formatted columns to a tidy results table, creating publication-ready output with properly formatted estimates, confidence intervals, and p-values.
Usage
format_results_table(
data,
est_col = "est",
lci_col = "lci",
uci_col = "uci",
pval_col = "pval",
est_digits = 2,
pval_digits = 3,
pval_threshold = 0.001,
ci_sep = ", "
)
Arguments
data |
A data.frame or tibble, typically output from |
est_col |
Character. Name of the estimate column. Default is "est". |
lci_col |
Character. Name of the lower CI column. Default is "lci". |
uci_col |
Character. Name of the upper CI column. Default is "uci". |
pval_col |
Character. Name of the p-value column. Default is "pval". |
est_digits |
Integer. Decimal places for estimates. Default is 2. |
pval_digits |
Integer. Decimal places for p-values. Default is 3. |
pval_threshold |
Numeric. Threshold for p-value formatting. Default is 0.001. |
ci_sep |
Character. Separator for CI bounds. Default is ", ". |
Details
This function is designed to work with output from tidy_pool_obj() but
can be used with any data.frame containing estimate, CI, and p-value columns.
The original columns are preserved; new formatted columns are added.
Value
A tibble with additional formatted columns:
- est_ci
Formatted estimate with confidence interval
- pval_fmt
Formatted p-value
Examples
library(dplyr)
# Create example results
results <- tibble::tibble(
parameter = c("trt_Week24", "lsm_ref_Week24", "lsm_alt_Week24"),
description = c("Treatment Effect", "LS Mean (Reference)", "LS Mean (Treatment)"),
est = c(-2.45, 5.20, 2.75),
se = c(0.89, 0.65, 0.71),
lci = c(-4.20, 3.93, 1.36),
uci = c(-0.70, 6.47, 4.14),
pval = c(0.006, NA, NA)
)
# Format for publication
formatted <- format_results_table(results)
print(formatted[, c("description", "est_ci", "pval_fmt")])
Utility function for Generalized G-computation for Binary Outcomes
Description
Wrapper function for targeting a marginal treatment effect using g-computation using the beeca package. Intended for binary endpoints.
Usage
gcomp_binary(
data,
outcome = "CRIT1FLN",
treatment = "TRT",
covariates = c("BASE", "STRATA", "REGION"),
reference = "Placebo",
contrast = "diff",
method = "Ge",
type = "HC0",
...
)
Arguments
data |
A data.frame containing the analysis dataset. |
outcome |
Name of the binary outcome variable (as string). |
treatment |
Name of the treatment variable (as string). |
covariates |
Character vector of covariate names to adjust for. |
reference |
Reference level for the treatment variable (default: "Placebo"). |
contrast |
Type of contrast to compute (default: "diff"). |
method |
Marginal estimation method for variance (default: "Ge"). |
type |
Variance estimator type (default: "HC0"). |
... |
Additional arguments passed to |
Value
A named list with treatment effect estimate, standard error, and degrees of freedom (if applicable).
Examples
# Load required packages
library(rbmiUtils)
library(beeca) # for get_marginal_effect()
library(dplyr)
# Load example data
data("ADMI")
# Ensure correct factor levels
ADMI <- ADMI |>
mutate(
TRT = factor(TRT, levels = c("Placebo", "Drug A")),
STRATA = factor(STRATA),
REGION = factor(REGION)
)
# Apply g-computation for binary responder
result <- gcomp_binary(
data = ADMI,
outcome = "CRIT1FLN",
treatment = "TRT",
covariates = c("BASE", "STRATA", "REGION"),
reference = "Placebo",
contrast = "diff",
method = "Ge", # from beeca: GEE robust sandwich estimator
type = "HC0" # from beeca: heteroskedasticity-consistent SE
)
# Print results
print(result)
G-computation Analysis for a Single Visit
Description
Performs logistic regression and estimates marginal effects for binary outcomes.
Usage
gcomp_responder(
data,
vars,
reference_levels = NULL,
var_method = "Ge",
type = "HC0",
contrast = "diff"
)
Arguments
data |
A data.frame with one visit of data. |
vars |
A list containing |
reference_levels |
Optional vector specifying reference level(s) of the treatment factor. |
var_method |
Marginal variance estimation method (default: "Ge"). |
type |
Type of robust variance estimator (default: "HC0"). |
contrast |
Type of contrast to compute (default: "diff"). |
Value
A named list containing estimates and standard errors for treatment comparisons and within-arm means.
Examples
library(dplyr)
library(rbmi)
library(rbmiUtils)
data("ADMI")
# Prepare data for a single visit
ADMI <- ADMI |>
mutate(
TRT = factor(TRT, levels = c("Placebo", "Drug A")),
STRATA = factor(STRATA),
REGION = factor(REGION)
)
dat_single <- ADMI |>
filter(AVISIT == "Week 24")
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CRIT1FLN",
covariates = c("BASE", "STRATA", "REGION")
)
result <- gcomp_responder(
data = dat_single,
vars = vars,
reference_levels = "Placebo"
)
print(result)
G-computation for a Binary Outcome at Multiple Visits
Description
Applies gcomp_responder() separately for each unique visit in the data.
Usage
gcomp_responder_multi(data, vars, reference_levels = NULL, ...)
Arguments
data |
A data.frame containing multiple visits. |
vars |
A list specifying analysis variables. |
reference_levels |
Optional reference level for the treatment variable. |
... |
Additional arguments passed to |
Value
A named list of estimates for each visit and treatment group.
Examples
library(dplyr)
library(rbmi)
library(rbmiUtils)
data("ADMI")
ADMI <- ADMI |>
mutate(
TRT = factor(TRT, levels = c("Placebo", "Drug A")),
STRATA = factor(STRATA),
REGION = factor(REGION)
)
# Note: method must match the original used for imputation
method <- method_bayes(
n_samples = 100,
control = control_bayes(warmup = 20, thin = 2)
)
vars_binary <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CRIT1FLN",
covariates = c("BASE", "STRATA", "REGION")
)
ana_obj_prop <- analyse_mi_data(
data = ADMI,
vars = vars_binary,
method = method,
fun = gcomp_responder_multi,
reference_levels = "Placebo",
contrast = "diff",
var_method = "Ge",
type = "HC0"
)
pool(ana_obj_prop)
Get Imputed Data Sets as a data frame
Description
This function takes an imputed dataset and a mapping variable to return a dataset with the original IDs mapped back and renamed appropriately.
Usage
get_imputed_data(impute_obj)
Arguments
impute_obj |
The imputation object from which the imputed datasets are extracted. |
Value
A data frame with the original subject IDs mapped and renamed.
Examples
library(dplyr)
library(rbmi)
library(rbmiUtils)
set.seed(1974)
# Load example dataset
data("ADEFF")
# Prepare data
ADEFF <- ADEFF |>
mutate(
TRT = factor(TRT01P, levels = c("Placebo", "Drug A")),
USUBJID = factor(USUBJID),
AVISIT = factor(AVISIT)
)
# Define variables for imputation
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CHG",
covariates = c("BASE", "STRATA", "REGION")
)
# Define Bayesian imputation method
method <- method_bayes(
n_samples = 100,
control = control_bayes(warmup = 200, thin = 2)
)
# Generate draws and perform imputation
draws_obj <- draws(data = ADEFF, vars = vars, method = method)
impute_obj <- impute(draws_obj,
references = c("Placebo" = "Placebo", "Drug A" = "Placebo"))
# Extract imputed data with original subject IDs
admi <- get_imputed_data(impute_obj)
head(admi)
Create a Forest Plot from an rbmi Pool Object
Description
Takes an rbmi pool object and produces a publication-quality, three-panel forest plot using ggplot2 and patchwork. The plot displays treatment effect point estimates with confidence interval whiskers, an aligned table panel showing formatted estimates, and a p-value panel.
Usage
plot_forest(
pool_obj,
display = c("trt", "lsm"),
ref_value = NULL,
ci_level = NULL,
arm_labels = NULL,
title = NULL,
text_size = 3.5,
point_size = 3.5,
show_pvalues = TRUE,
font_family = NULL,
panel_widths = NULL
)
Arguments
pool_obj |
A pooled analysis object of class |
display |
Character string specifying the display mode. |
ref_value |
Numeric. The reference value for the vertical reference
line. Default is |
ci_level |
Numeric. Confidence level for CI labeling. If |
arm_labels |
Named character vector with elements |
title |
Optional character string for the plot title. |
text_size |
Numeric. Text size for the table and p-value panels. Default is 3.5. |
point_size |
Numeric. Point size for the forest plot. Default is 3.5. |
show_pvalues |
Logical. Whether to display the p-value panel on the
right side of the plot. Default is |
font_family |
Optional character string specifying the font family for
all text in the plot. When |
panel_widths |
Optional numeric vector controlling the relative widths
of the plot panels. When |
Details
The function calls tidy_pool_obj() internally to parse the pool object,
then constructs a three-panel composition:
-
Left panel: Visit labels and formatted estimate with CI text
-
Middle panel: Forest plot with point estimates and CI whiskers
-
Right panel: Formatted p-values
Display modes:
-
"trt"– Treatment differences with a reference line at zero (or customref_value). Significant results (CI excludes reference) are shown as filled circles; non-significant as open circles. -
"lsm"– LS mean estimates by treatment arm, color-coded using the Okabe-Ito colorblind-friendly palette (blue for reference, vermilion for treatment). Points are dodged vertically within each visit.
Customization: The returned patchwork object supports & theme() for
applying theme changes to all panels. For example:
plot_forest(pool_obj) & theme(text = element_text(size = 14)).
Suggested dimensions for regulatory documents: For A4 or US Letter page
sizes, width = 10, height = 3 + 0.4 * n_visits (in inches) provides good
results when saving with ggplot2::ggsave(). For example, a 5-visit plot
works well at 10 x 5 inches.
Example output (treatment difference mode):
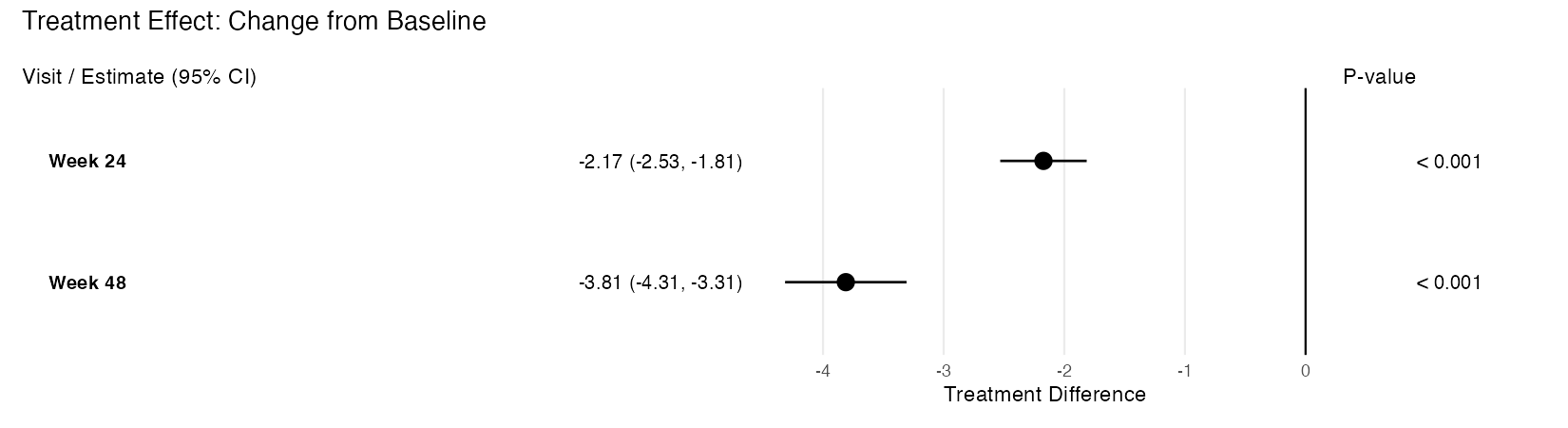
Value
A patchwork/ggplot object that can be further customized using
& theme() to modify all panels simultaneously.
See Also
-
rbmi::pool()for creating pool objects -
tidy_pool_obj()for the underlying data transformation -
efficacy_table()for tabular presentation of the same data -
format_pvalue()for p-value formatting rules -
format_estimate()for estimate with CI formatting
Examples
if (requireNamespace("ggplot2", quietly = TRUE) &&
requireNamespace("patchwork", quietly = TRUE)) {
library(rbmi)
data("ADMI", package = "rbmiUtils")
ADMI$TRT <- factor(ADMI$TRT, levels = c("Placebo", "Drug A"))
ADMI$USUBJID <- factor(ADMI$USUBJID)
ADMI$AVISIT <- factor(ADMI$AVISIT)
vars <- set_vars(
subjid = "USUBJID", visit = "AVISIT", group = "TRT",
outcome = "CHG", covariates = c("BASE", "STRATA", "REGION")
)
method <- method_bayes(
n_samples = 20,
control = control_bayes(warmup = 20, thin = 1)
)
ana_obj <- analyse_mi_data(ADMI, vars, method, fun = ancova)
pool_obj <- pool(ana_obj)
# Treatment difference forest plot
plot_forest(pool_obj, arm_labels = c(ref = "Placebo", alt = "Drug A"))
# LSM display with custom panel widths
plot_forest(
pool_obj,
display = "lsm",
arm_labels = c(ref = "Placebo", alt = "Drug A"),
title = "LS Mean Estimates by Visit",
panel_widths = c(3, 5, 1.5)
)
}
Convert Pool Object to ARD Format
Description
Converts an rbmi pool object to the pharmaverse Analysis Results Dataset (ARD) standard using the cards package. The ARD format is a long-format data frame where each row represents a single statistic for a given parameter, with grouping columns for visit, parameter type, and least-squares-mean type.
Usage
pool_to_ard(pool_obj, analysis_obj = NULL, conf.level = NULL)
Arguments
pool_obj |
A pooled analysis object of class |
analysis_obj |
An optional analysis object (output of
|
conf.level |
Confidence level used for CI labels (e.g., |
Details
The function works by:
Tidying the pool object via
tidy_pool_obj()Reshaping each parameter into long-format ARD rows (one row per statistic)
Adding grouping columns (visit, parameter_type, lsm_type)
Optionally enriching with MI diagnostic statistics when
analysis_objis providedApplying
cards::as_card()andcards::tidy_ard_column_order()for standard ARD structure
When analysis_obj is provided:
For Rubin's rules pooling: diagnostic statistics (FMI, lambda, RIV, Barnard-Rubin adjusted df, relative efficiency) are computed per parameter using the per-imputation estimates, standard errors, and degrees of freedom.
For non-Rubin pooling methods: an informative message is emitted and the base ARD is returned without diagnostic rows.
The resulting ARD passes cards::check_ard_structure() validation and is
suitable for downstream use with gtsummary.
Value
A data frame of class "card" (ARD format) with grouping columns
for visit (group1), parameter_type (group2), and lsm_type (group3).
Each parameter produces rows for five statistics: estimate, std.error,
conf.low, conf.high, and p.value, plus a method row. When analysis_obj
is provided and the pooling method is Rubin's rules, additional diagnostic
stat rows are included: fmi, lambda, riv, df.adjusted, df.complete, re,
and m.imputations.
See Also
-
rbmi::pool()for creating pool objects -
analyse_mi_data()for creating analysis objects -
tidy_pool_obj()for the underlying data transformation -
cards::as_card()andcards::check_ard_structure()for ARD validation
Examples
if (requireNamespace("cards", quietly = TRUE)) {
library(rbmi)
data("ADMI", package = "rbmiUtils")
ADMI$TRT <- factor(ADMI$TRT, levels = c("Placebo", "Drug A"))
ADMI$USUBJID <- factor(ADMI$USUBJID)
ADMI$AVISIT <- factor(ADMI$AVISIT)
vars <- set_vars(
subjid = "USUBJID", visit = "AVISIT", group = "TRT",
outcome = "CHG", covariates = c("BASE", "STRATA", "REGION")
)
method <- method_bayes(
n_samples = 20,
control = control_bayes(warmup = 20, thin = 1)
)
ana_obj <- analyse_mi_data(ADMI, vars, method, fun = ancova)
pool_obj <- pool(ana_obj)
# Base ARD
ard <- pool_to_ard(pool_obj)
# Enriched ARD with MI diagnostics (FMI, lambda, RIV, df)
ard_diag <- pool_to_ard(pool_obj, analysis_obj = ana_obj)
}
Prepare Intercurrent Event Data
Description
Builds a data_ice data.frame from a column in the dataset that flags
intercurrent events. For each subject, the first visit (by factor level
order) where the flag is TRUE is used as the ICE visit.
Usage
prepare_data_ice(data, vars, ice_col, strategy)
Arguments
data |
A data.frame containing the analysis dataset. |
vars |
A |
ice_col |
Character string naming the column in |
strategy |
Character string specifying the imputation strategy to assign.
Must be one of |
Value
A data.frame with columns corresponding to vars$subjid,
vars$visit, and vars$strategy, suitable for passing to
rbmi::draws().
See Also
-
rbmi::draws()which accepts thedata_iceoutput from this function -
validate_data()to check data before imputation -
summarise_missingness()to understand missing data patterns
Examples
library(rbmi)
dat <- data.frame(
USUBJID = factor(rep(c("S1", "S2", "S3"), each = 3)),
AVISIT = factor(rep(c("Week 4", "Week 8", "Week 12"), 3),
levels = c("Week 4", "Week 8", "Week 12")),
TRT = factor(rep(c("Placebo", "Drug A", "Drug A"), each = 3)),
CHG = rnorm(9),
DISCFL = c("N","N","N", "N","Y","Y", "N","N","Y")
)
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CHG"
)
ice <- prepare_data_ice(dat, vars, ice_col = "DISCFL", strategy = "JR")
print(ice)
Print Method for Analysis Objects
Description
Prints a summary of an analysis object from analyse_mi_data().
Usage
## S3 method for class 'analysis'
print(x, ...)
Arguments
x |
An object of class |
... |
Additional arguments (currently unused). |
Value
Invisibly returns the input object.
Examples
library(rbmi)
library(rbmiUtils)
data("ADMI")
# Create analysis object
vars <- set_vars(
subjid = "USUBJID", visit = "AVISIT", group = "TRT",
outcome = "CHG", covariates = c("BASE", "STRATA")
)
method <- method_bayes(n_samples = 10, control = control_bayes(warmup = 10))
ana_obj <- analyse_mi_data(ADMI, vars, method, fun = function(d, v, ...) 1)
print(ana_obj)
Print Method for describe_draws Objects
Description
Displays a formatted summary of a draws description using cli formatting.
Usage
## S3 method for class 'describe_draws'
print(x, ...)
Arguments
x |
A |
... |
Additional arguments (currently unused). |
Value
Invisibly returns x (for pipe chaining).
Examples
## Not run:
# After creating draws_obj via the rbmi pipeline (see describe_draws):
desc <- describe_draws(draws_obj)
print(desc) # Formatted cli output with method, formula, samples, convergence
## End(Not run)
Print Method for describe_imputation Objects
Description
Displays a formatted summary of an imputation description using cli formatting, including method, number of imputations, reference arm mappings, and a missingness breakdown by visit and treatment arm.
Usage
## S3 method for class 'describe_imputation'
print(x, ...)
Arguments
x |
A |
... |
Additional arguments (currently unused). |
Value
Invisibly returns x (for pipe chaining).
Examples
## Not run:
# After creating impute_obj via the rbmi pipeline (see describe_imputation):
desc <- describe_imputation(impute_obj)
print(desc) # Formatted cli output with method, M, subjects, references, missingness
## End(Not run)
Print Method for Pool Objects
Description
Displays a formatted summary of a pooled analysis object from rbmi::pool().
Uses cli formatting to show rounded estimates, confidence intervals, parameter
labels, method information, number of imputations, and confidence level.
Usage
## S3 method for class 'pool'
print(x, digits = 2, ...)
Arguments
x |
An object of class |
digits |
Integer. Number of decimal places for rounding estimates, standard errors, and confidence interval bounds. Default is 2. |
... |
Additional arguments (currently unused). |
Details
This method overrides rbmi::print.pool() to provide enhanced, formatted
console output using the cli package. The override produces a
"Registered S3 method overwritten" message at package load time, which is
expected and harmless (same pattern as print.analysis()).
The output includes:
A header with parameter and visit counts
Metadata: pooling method, number of imputations, confidence level
A compact results table with key columns: parameter, visit, est, lci, uci, pval
Value
Invisibly returns the original pool object x (for pipe chaining).
See Also
-
tidy_pool_obj()for full tidy tibble output -
summary.pool()for visit-level breakdown with significance flags -
rbmi::pool()to create pool objects
Examples
library(rbmi)
library(rbmiUtils)
data("ADMI")
ADMI$TRT <- factor(ADMI$TRT, levels = c("Placebo", "Drug A"))
ADMI$USUBJID <- factor(ADMI$USUBJID)
ADMI$AVISIT <- factor(ADMI$AVISIT)
vars <- set_vars(
subjid = "USUBJID", visit = "AVISIT", group = "TRT",
outcome = "CHG", covariates = c("BASE", "STRATA", "REGION")
)
method <- method_bayes(n_samples = 20, control = control_bayes(warmup = 20))
ana_obj <- analyse_mi_data(ADMI, vars, method, fun = ancova)
pool_obj <- pool(ana_obj)
print(pool_obj)
Reduce Imputed Data for Efficient Storage
Description
Extracts only the imputed records (those that were originally missing) from a full imputed dataset. This significantly reduces storage requirements when working with many imputations, as observed values are identical across all imputations and only need to be stored once in the original data.
Usage
reduce_imputed_data(imputed_data, original_data, vars)
Arguments
imputed_data |
A data.frame containing the full imputed dataset with an
|
original_data |
A data.frame containing the original dataset before imputation, with missing values in the outcome column. |
vars |
A |
Details
Storage savings depend on the proportion of missing data. For example:
Original: 1000 rows, 44 missing values
Full imputed (1000 imputations): 1,000,000 rows
Reduced (1000 imputations): 44,000 rows (4.4\
Use expand_imputed_data() to reconstruct the full imputed dataset when
needed for analysis.
Value
A data.frame containing only the rows from imputed_data that
correspond to originally missing outcome values. All columns from
imputed_data are preserved.
See Also
-
rbmi::impute()which creates the imputed datasets this function operates on -
expand_imputed_data()to reconstruct the full dataset -
get_imputed_data()to extract imputed data from an rbmi imputation object
Examples
library(rbmi)
library(dplyr)
# Example with package data
data("ADMI", package = "rbmiUtils")
data("ADEFF", package = "rbmiUtils")
# Prepare original data to match ADMI structure
original <- ADEFF |>
mutate(
TRT = TRT01P,
USUBJID = as.character(USUBJID)
)
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CHG"
)
# Reduce to only imputed values
reduced <- reduce_imputed_data(ADMI, original, vars)
# Compare sizes
cat("Full imputed rows:", nrow(ADMI), "\n")
cat("Reduced rows:", nrow(reduced), "\n")
cat("Compression:", round(100 * nrow(reduced) / nrow(ADMI), 1), "%\n")
Summarise Missing Data Patterns
Description
Tabulates missing outcome data by visit and treatment group, and classifies each subject's missing data pattern as complete, monotone, or intermittent.
Usage
summarise_missingness(data, vars)
Arguments
data |
A data.frame containing the analysis dataset with one row per subject-visit combination. |
vars |
A |
Value
A list with three components:
- by_visit
A tibble with columns: visit, group, n, n_miss, pct_miss
- patterns
A tibble with columns: subjid, group, pattern ("complete", "monotone", or "intermittent"), dropout_visit (NA if not monotone)
- summary
A tibble with columns: group, n_subjects, n_complete, n_monotone, n_intermittent
See Also
-
rbmi::draws()for imputation after reviewing missingness patterns -
validate_data()to check data before imputation -
prepare_data_ice()to create intercurrent event data from flags
Examples
library(rbmi)
dat <- data.frame(
USUBJID = factor(rep(c("S1", "S2", "S3", "S4"), each = 3)),
AVISIT = factor(rep(c("Week 4", "Week 8", "Week 12"), 4),
levels = c("Week 4", "Week 8", "Week 12")),
TRT = factor(rep(c("Placebo", "Drug A"), each = 6)),
CHG = c(1, 2, 3, 1, NA, NA, 1, 2, NA, 1, NA, 2)
)
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CHG"
)
result <- summarise_missingness(dat, vars)
print(result$by_visit)
print(result$patterns)
print(result$summary)
Summary Method for Analysis Objects
Description
Provides a detailed summary of an analysis object from analyse_mi_data().
Usage
## S3 method for class 'analysis'
summary(object, n_preview = 5, ...)
Arguments
object |
An object of class |
n_preview |
Maximum number of parameters to show in the preview table. Defaults to 5. |
... |
Additional arguments (currently unused). |
Value
A list containing summary information (invisibly).
Examples
library(rbmi)
library(rbmiUtils)
data("ADMI")
# Create analysis object
vars <- set_vars(
subjid = "USUBJID", visit = "AVISIT", group = "TRT",
outcome = "CHG", covariates = c("BASE", "STRATA")
)
method <- method_bayes(n_samples = 10, control = control_bayes(warmup = 10))
ana_obj <- analyse_mi_data(ADMI, vars, method, fun = function(d, v, ...) 1)
summary(ana_obj)
Summary Method for Pool Objects
Description
Provides a detailed visit-level breakdown of pooled analysis results with significance flags. Shows treatment comparisons and least squares means grouped by visit.
Usage
## S3 method for class 'pool'
summary(object, alpha = 0.05, ...)
Arguments
object |
An object of class |
alpha |
Numeric. Significance threshold for flagging p-values.
Default is 0.05. Flags are: |
... |
Additional arguments (currently unused). |
Details
The summary output groups results by visit, showing treatment comparisons with significance flags and least squares means. This provides a quick overview of which visits have statistically significant treatment effects.
Significance flags:
-
*p < alpha (default 0.05) -
**p < 0.01 -
***p < 0.001
Value
Invisibly returns a list with:
- n_parameters
Number of parameters in the pool object
- visits
Character vector of unique visit names
- method
Pooling method used
- n_imputations
Number of imputations combined
- conf.level
Confidence level
- tidy_df
The full tidy tibble from
tidy_pool_obj()
See Also
-
print.pool()for compact tabular output -
tidy_pool_obj()for full tidy tibble output -
rbmi::pool()to create pool objects
Examples
library(rbmi)
library(rbmiUtils)
data("ADMI")
ADMI$TRT <- factor(ADMI$TRT, levels = c("Placebo", "Drug A"))
ADMI$USUBJID <- factor(ADMI$USUBJID)
ADMI$AVISIT <- factor(ADMI$AVISIT)
vars <- set_vars(
subjid = "USUBJID", visit = "AVISIT", group = "TRT",
outcome = "CHG", covariates = c("BASE", "STRATA", "REGION")
)
method <- method_bayes(n_samples = 20, control = control_bayes(warmup = 20))
ana_obj <- analyse_mi_data(ADMI, vars, method, fun = ancova)
pool_obj <- pool(ana_obj)
summary(pool_obj)
Tidy and Annotate a Pooled Object for Publication
Description
This function processes a pooled analysis object of class pool into a tidy tibble format.
It adds contextual information, such as whether a parameter is a treatment comparison or a least squares mean,
dynamically identifies visit names from the parameter column, and provides additional columns for parameter type,
least squares mean type, and visit.
Usage
tidy_pool_obj(pool_obj)
Arguments
pool_obj |
A pooled analysis object of class |
Details
The function dynamically processes the parameter column by separating it into
components (e.g., type of estimate, reference vs. alternative arm, and visit), and provides
informative descriptions in the output.
Workflow:
Prepare data and run imputation with rbmi
Analyse with
analyse_mi_data()Pool with
rbmi::pool()Tidy with
tidy_pool_obj()for publication-ready output
Value
A tibble containing the processed pooled analysis results with the following columns:
- parameter
Original parameter name from the pooled object
- description
Human-readable description of the parameter
- visit
Visit name extracted from parameter (if applicable)
- parameter_type
Either "trt" (treatment comparison) or "lsm" (least squares mean)
- lsm_type
For LSM parameters: "ref" (reference) or "alt" (alternative)
- est
Point estimate
- se
Standard error
- lci
Lower confidence interval
- uci
Upper confidence interval
- pval
P-value
See Also
-
rbmi::pool()which creates the pool objects this function tidies -
analyse_mi_data()to analyse imputed datasets -
format_results()for additional formatting options
Examples
# Example usage:
library(dplyr)
library(rbmi)
data("ADMI")
N_IMPUTATIONS <- 100
BURN_IN <- 200
BURN_BETWEEN <- 5
# Convert key columns to factors
ADMI$TRT <- factor(ADMI$TRT, levels = c("Placebo", "Drug A"))
ADMI$USUBJID <- factor(ADMI$USUBJID)
ADMI$AVISIT <- factor(ADMI$AVISIT)
# Define key variables for ANCOVA analysis
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CHG",
covariates = c("BASE", "STRATA", "REGION") # Covariates for adjustment
)
# Specify the imputation method (Bayesian) - need for pool step
method <- rbmi::method_bayes(
n_samples = N_IMPUTATIONS,
control = rbmi::control_bayes(
warmup = BURN_IN,
thin = BURN_BETWEEN
)
)
# Perform ANCOVA Analysis on Each Imputed Dataset
ana_obj_ancova <- analyse_mi_data(
data = ADMI,
vars = vars,
method = method,
fun = ancova, # Apply ANCOVA
delta = NULL # No sensitivity analysis adjustment
)
pool_obj_ancova <- pool(ana_obj_ancova)
tidy_df <- tidy_pool_obj(pool_obj_ancova)
# Print tidy data frames
print(tidy_df)
Validate Data Before Imputation
Description
Pre-flight validation of data, variable specification, and intercurrent event
data before calling rbmi::draws(). Collects all issues and reports them
together in a single error message.
Usage
validate_data(data, vars, data_ice = NULL)
Arguments
data |
A data.frame containing the analysis dataset. |
vars |
A |
data_ice |
An optional data.frame of intercurrent events. If provided,
must contain columns corresponding to |
Details
The following checks are performed:
-
datais a data.frame All columns named in
varsexist indata-
subjid,visit, andgroupcolumns are factors -
outcomecolumn is numeric Covariate columns have no missing values
Data has one row per subject-visit combination
If
data_iceis provided: correct columns, valid subjects, valid visits, recognised strategies, and at most one row per subject
Recommended Workflow:
Call
validate_data()to check your dataUse
prepare_data_ice()to create ICE data if neededReview missingness with
summarise_missingness()Proceed with
rbmi::draws()for imputation
Value
Invisibly returns TRUE if all checks pass. Throws an error with
collected messages if any issues are found.
See Also
-
rbmi::draws()which requires validated input data -
prepare_data_ice()to create intercurrent event data from flags -
summarise_missingness()to understand missing data patterns
Examples
library(rbmi)
dat <- data.frame(
USUBJID = factor(rep(c("S1", "S2", "S3"), each = 3)),
AVISIT = factor(rep(c("Week 4", "Week 8", "Week 12"), 3),
levels = c("Week 4", "Week 8", "Week 12")),
TRT = factor(rep(c("Placebo", "Drug A", "Drug A"), each = 3)),
CHG = c(1.1, 2.2, 3.3, 0.5, NA, NA, 1.0, 2.0, NA),
BASE = rep(c(10, 12, 11), each = 3),
STRATA = factor(rep(c("A", "B", "A"), each = 3))
)
vars <- set_vars(
subjid = "USUBJID",
visit = "AVISIT",
group = "TRT",
outcome = "CHG",
covariates = c("BASE", "STRATA")
)
validate_data(dat, vars)